Big Data in the Arts and Humanities
Big Data in the Arts and Humanities: Some Arts and Humanities Research Council Projects

Mining the History of Medicine
How can a technique for analysing ‘big data’ usually employed in biomedicine be used to explore the history of medicine? This is the question that brought together our University of Manchester teams, the National Centre for Text Mining (NaCTeM) and the Centre for the History of Science, Technology and Medicine (CHSTM), to collaborate on the Mining the History of Medicine project. The historical data we worked with – full-text collections of the British Medical Journal (BMJ) (1840-present) and the Medical Officer of Health reports from the London boroughs (MOH) (1848-1972) – although relatively small by conventional ‘big data’ standards, can nevertheless be overwhelming in scale, even for experienced historical researchers. We thus decided to use text mining (TM) techniques to enhance and expand the possibilities for using these resources. Firstly, we set out to make exploring these vast repositories of past medical information faster and more efficient. Secondly, we wanted to allow these materials to be used in new ways, as well making them accessible to a wider variety of scholars, ranging from other historians, to environmental scholars, to contemporary biomedical researchers. By developing a semantic search engine (http://www.nactem.ac.uk/hom/), underpinned by sophisticated TM techniques, we would allow historians and other scholars to see the content of the BMJ and MOH in new ways, by keeping both the big picture and the fine detail in view at the same time.
Our collaboration has produced a unique and useful resource about health and medicine in Britain’s past. Our first contribution was to develop a method to correct OCR errors, based on a spellchecking approach adapted to the medical domain. Given that the BMJ’s original OCR text had a word error rate of up to 30%, correcting as many OCR errors as possible was important to ensure that TM tools performed accurately. Secondly, the historians worked with the TM specialists to adapt TM tools to automatically recognise semantic entities (e.g., medical conditions, signs/symptoms, therapeutic measures, environmental factors etc.). This was an exciting challenge for both parts of the team: was it possible to develop TM tools that could robustly recognise historically and medically appropriate entities and relationships between them, in a time-sensitive manner? After all, we wanted to teach the system to detect discussions about disease and its causes, at the very time that the concept of disease and understanding of its causation was being radically redefined.
Our search system makes it possible to discover relevant documents from different periods of time. Entering pulmonary tuberculosis will result in the suggestion of related terms, such as pulmonary phthisis, which may be unknown to non-experts, but which can allow the retrieval of older relevant documents. Graphs show how the frequency of these terms changes over time, which can reveal, e.g., when the condition became controllable.
Rather than being presented simply with a list of thousands of documents, users can easily explore the semantic content of search results, e.g., to discover which types of environmental factors are mentioned in the context of tuberculosis. These range from foodstuffs such as meat, to issues such as overcrowding and hygiene. Documents containing entities of interest may be isolated, making it easier to focus on particular topics, or to explore (potentially unknown) associations between entities. Moving a step further, the system can allow temporal tracking of answers to specific questions such as ‘What causes tuberculosis?’
Positive and constructive feedback received during a recent workshop Expert Advisory User Group has reinforced the success of applying TM techniques to historical medical text, and demonstrates the potential of developing innovative TM tools for a wider range of historical text types.
Research team: University of Manchester, National Centre for Text Mining: Sophia Ananiadou, John McNaught, Carsten Timmermann, Paul Thompson, Jacob Carter; University of Manchester, Centre for the History of Science, Technology and Medicine: Michael Worboys; Elizabeth Toon
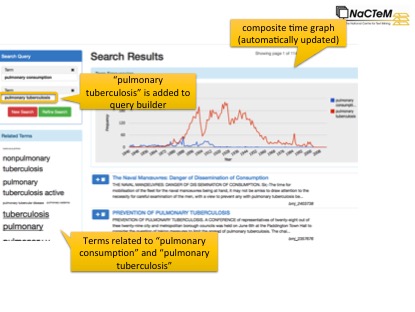
Image: Search interface from Mining the History of Medicine. Searching fora term will: display documents containing the term; dispaly a graph showing the term’s frequency over time; and show other terms semantically related to the search term (here we see several other repository conditions). Clicking on a term in the ‘Related terms’ box causes it to be added to the query. Documents and related terms are updated accordingly, and the graph is updated to allow the historical paths of terms to be compared.
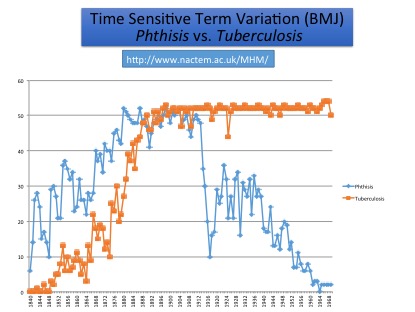
Mining the History of Medicine: in the 19th century, the term ‘Phthisis’ was preferred to ‘Tuberculosis’, but its use declined in the 20th century. In searching, it is vital to remember the earlier preference for ‘Phthisis’, otherwise references will be missed